1.9 Working with Large Datasets
The learning objectives of this section are to:
- Read and manipulate large datasets
R now offers now offers a variety of options for working with large datasets. We won’t try to cover all these options in detail here, but rather give an overview of strategies to consider if you need to work with a large dataset, as well as point you to additional resources to learn more about working with large datasets in R.
A> While there are a variety of definitions of how large a dataset must be to qualify as “large,” in this section we don’t formally define a limit. Instead, this section is meant to give you some strategies anytime you work with a dataset large enough that you notice it’s causing problems. For example, data large enough for R to be noticeably slow to read or manipulate the data, or large enough it’s difficult to store the data locally on your computer.
1.9.1 In-memory strategies
A> In this section, we introduce the basics of why and how to use data.table to work with large datasets in R. We have included a video demonstration online showing how functions from the data.table package can be used to load and explore a large dataset more efficiently.
The data.table package can help you read a large dataset into R and explore it more efficiently. The fread function in this package, for example, can read large flat files in much more quickly than comparable base R packages. Since all of the data.table functions will work with smaller datasets, as well, we’ll illustrate using data.table with the Zika data accessed from GitHub in an earlier section of this chapter. We’ve saved that data locally to illustrate how to read it in and work with it using data.table.
First, to read this data in using fread, you can run:
library(data.table)
brazil_zika <- fread("data/COES_Microcephaly-2016-06-25.csv")
head(brazil_zika, 2)
report_date location location_type data_field
1: 2016-06-25 Brazil-Acre state microcephaly_confirmed
2: 2016-06-25 Brazil-Alagoas state microcephaly_confirmed
data_field_code time_period time_period_type value unit
1: BR0002 NA NA 2 cases
2: BR0002 NA NA 75 cases
class(brazil_zika)
[1] "data.table" "data.frame"If you are working with a very large dataset, data.table will provide a status bar showing your progress towards loading the code as you read it in using fread.
If you have a large dataset for which you only want to read in certain columns, you can save time when using data.table by only reading in the columns you want with the select argument in fread. This argument takes a vector of either the names or positions of the columns that you want to read in:
fread("data/COES_Microcephaly-2016-06-25.csv",
select = c("location", "value", "unit")) %>%
dplyr::slice(1:3)
location value unit
1: Brazil-Acre 2 cases
2: Brazil-Alagoas 75 cases
3: Brazil-Amapa 7 casesMany of the fread arguments are counterparts to arguments in the read.table family of functions in base R (for example, na.strings, sep, skip, colClasses). One that is particular useful is nrows. If you’re working with data that takes a while to read in, using nrows = 20 or some other small number will allow you to make sure you have set all of the arguments in fread appropriately for the dataset before you read in the full dataset.
If you already have a dataset loaded to your R session, you can use the data.table function to convert a data frame into a data.table object. (Note: if you use fread, the data is automatically read into a data.table object.) A data.table object also has the class data.frame; this means that you can use all of your usual methods for manipulating a data frame with a data.table object. However, for extra speed, use data.table functions to manipulate, clean, and explore the data in a data.table object. You can find out more about using data.table functions at the data.table wiki.
A> Many of the functions in data.table, like many in ddplyr, use non-standard evaluation. This means that, while they’ll work fine in interactive programming, you’ll need to take some extra steps when you use them to write functions for packages. We’ll cover non-standard evaluation in the context of developing packages in a later section.
When you are working with datasets that are large, but can still fit in-memory, you’ll want to optimize your code as much as possible. There are more details on profiling and optimizing code in a later chapter, but one strategy for speeding up R code is to write some of the code in C++ and connect it to R using the Rcpp package. Since C++ is a compiled rather than an interpreted language, it runs much faster than similar code written in R. If you are more comfortable coding in another compiled language (C or FORTRAN, for example), you can also use those, although the Rcpp package is very nicely written and well-maintained, which makes C++ an excellent first choice for creating compiled code to speed up R.
Further, a variety of R packages have been written that help you run R code in parallel, either locally or on a cluster. Parallel strategies may be work pursuing if you are working with very large datasets, and if the coding tasks can be split to run in parallel. To get more ideas and find relevant packages, visit CRAN’s High-Performance and Parallel Computing with R task view.
1.9.2 Out-of-memory strategies
If you need to work with a very large dataset, there are also some options to explore and model the dataset without ever loading it into R, while still using R commands and working from the R console or an R script. These options can make working with large datasets more efficient, because they let other software handle the heavy lifting of sifting through the data and / or avoid loading large datasets into RAM, instead using data stored on hard drive.
For example, database management systems are optimized to more efficiently store and better search through large sets of data; popular examples include Oracle, MySQL, and PostgreSQL. There are several R packages that allow you to connect your R session to a database. With these packages, you can use functions from the R console or an R script to search and subset data without loading the whole dataset into R, and so take advantage of the improved efficiency of the database management system in handling data, as well as work with data too big to fit in memory.
The DBI package is particularly convenient for interfacing R code with a database management system, as it provides a top-level interface to a number of different database management systems, with system-specific code applied by a lower-level, more specific R package (Figure 1.7).
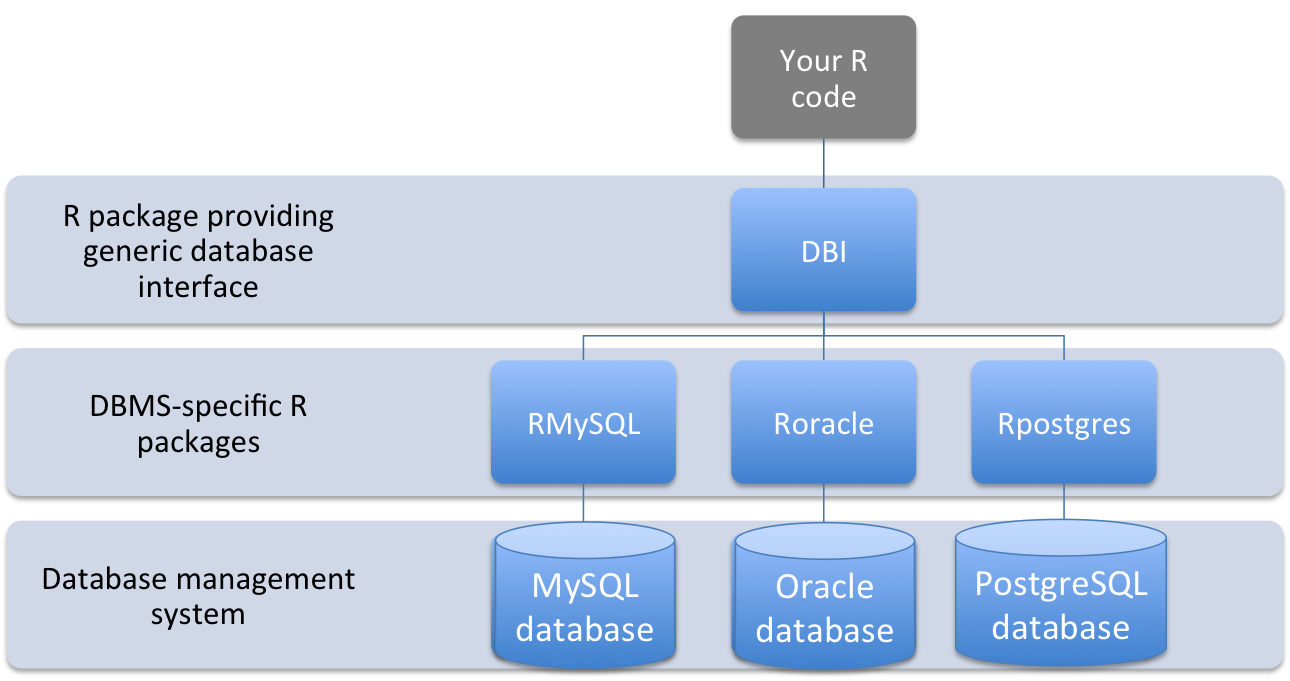
Figure 1.7: Structure of interface between code in an R script and data stored in a database management system using DBI-compliant packages
The DBI package therefore allows you to use the same commands for working with database-stored data in R, without worrying about details specific to the exact type of database management system you’re connecting to. The following table outlines the DBI functions you can use to perform a variety of tasks when working with data stored in a database:
| Task | DBI Function |
|---|---|
| Create a new driver object for an instance of a database | dbDriver |
| Connect to database instance | dbConnect |
| Find available tables in a connected database instance | dbListTables |
| Find available fields within a table | dbListFields |
| Query a connected database instance | dbSendQuery |
| Pull a data frame into R from a query result | dbFetch |
| Jointly query and pull data from a database instance | dbGetQuery |
| Close result set from a query | dbClearResult |
| Write a new table in a database instance | dbWriteTable |
| Remove a table from a database instance | dbRemoveTable |
| Disconnect from a database instance | dbDisconnect |
The DBI package depends on lower-level R packages to translate its generic commands to work for specific database management systems. DBI-compliant R packages have not been written for every database management system, so there are some databases for which DBI commands will not work. DBI-compliant R packages that are available include:
| Database Management System | R packages |
|---|---|
| Oracle | ROracle |
| MySQL | RMySQL |
| Microsoft SQL Server | RSQLServer |
| PostgreSQL | RPostgres |
| SQLite | RSQLite |
I> For more on the DBI package, including its history, see the package’s GitHub README page.
The packages for working with database management systems require you to send commands to the database management system in that system’s command syntax (e.g., SQL). You can, however, do “SELECT” database queries directly using dplyr syntax for some database systems, rather than with SQL syntax. While this functionality is limited to “SELECT” calls, often this is all you’ll need within a data analysis script. For more details, see the dplyr database vignette.
In addition to database management systems, there are other options for working with large data out-of-memory in R. For example, the bigmemory and associated packages can be used to access and work with large matrices stored on hard drive rather than in RAM, by storing the data in a C++ matrix structure and loading to R pointers to the data, rather than the full dataset. This family of packages includes packages that can be used to summarize and model the data (biglm, bigglm, biganalytics, bigtabulate, bigalgebra). One limitation is that these packages only work with matrices, not data frames; matrices require all elements share a class (e.g., all numeric).
Finally, there are some packages that allow you to write R code that uses other software to load and work with data through an R API provided by the other software. For example, the h2o package allows you to write R code to load and fit machine learning models in H2O, which is open-source software that facilitates distributed machine learning. H2O includes functions to fit and evaluate numerous machine learning models, including ensemble models, which would take quite a while to fit within R with a large training dataset. Since processing is done using compiled code, models can be fit on large datasets more quickly. However, while the h2o package allows you to use R-like code from within an R console to explore and model your data, it is not actually running R, but instead is using the R code, through the R API, to run Java-encoded functions. As a result, you only have access to a small subset of R’s total functionality, since you can only run the R-like functions written into H2O’s own software.